Discovery of tsRNAs in mature sperm
Hidden information in sperm small RNAs: tRNA-derived small RNA (tsRNA) & rRNA-derived small RNA (rsRNA)
Discovery of tsRNAs in serum, across vertebrate species
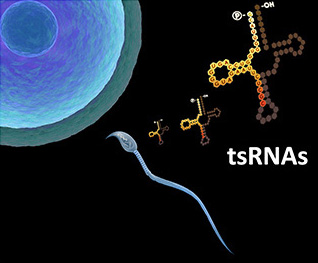
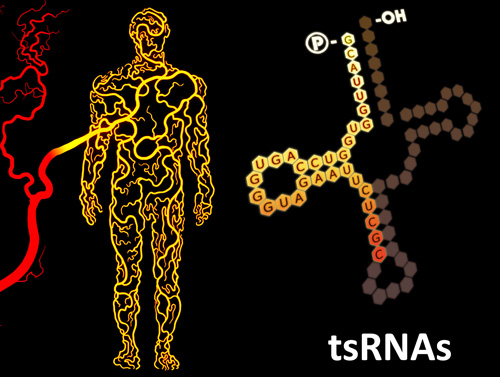
Sperm tsRNAs/rsRNAs, and RNA modifications, contribute to epigenetic inheritance of paternally acquired traits
Parallel with the discovery of tsRNAs in sperm and serum, we are fascinated about the increasing evidence that certain acquired traits during paternal exposure can be “memorized” in the sperm and inherited by the offspring, leading to a resurrection of the ideas of 'Lamarckian inheritance' and 'Darwin's Pangenesis' (Nat Rev Genet 2016, Nat Rev Mol Cell Biol 2018). Recently, we found changes in expression profiles in sperm tsRNAs of mice with a paternal high-fat diet (HFD) (Science 2016), and later on also found another type of small RNAs, rRNA-derived small RNAs (rsRNAs), co-exist with tsRNAs in the sperm and they are sensitively regulated by HFD (Nat Cell Biol 2018). By injecting sperm total RNAs or different RNA fractions into normal zygotes, we found that the tsRNA/rsRNA-enriched 30-40nt sperm RNA fraction, along with associated RNA modifications, represent a carrier of paternal epigenetic information that contributes to intergenerational inheritance of diet-induced metabolic disorder (Science 2016; Nat Cell Biol 2018). We also demonstrated that a RNA methyltransferase, Dnmt2, can shape the sperm RNA ‘coding signature’ by regulating RNA modifications and tsRNA/rsRNA biogenesis, and affect RNAs’ secondary structures that may together contribute to the effect of epigenetic inheritance (Nat Cell Biol 2018).
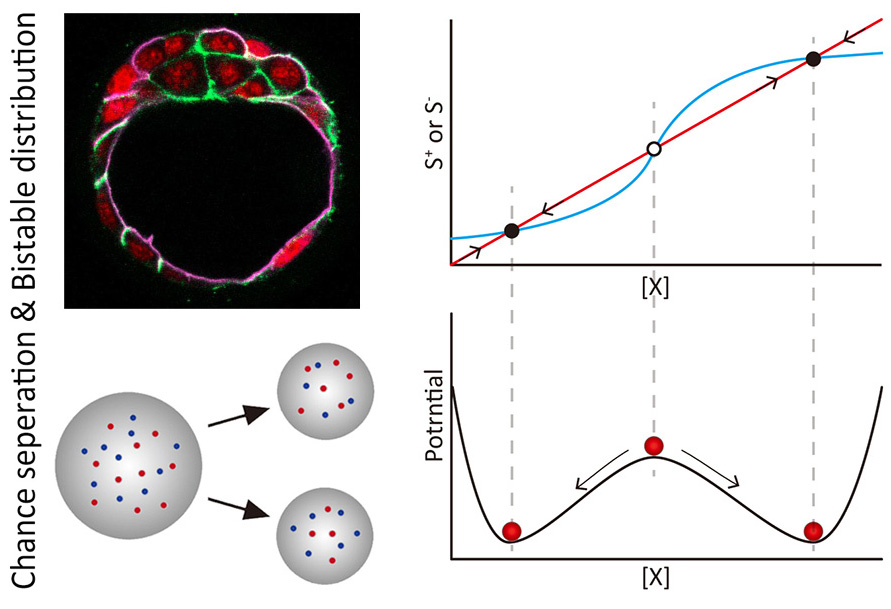

Single-cell analysis: bridging randomness and determinism

Compartmentalized intracellular reactions: an alternative source of heterogeneity
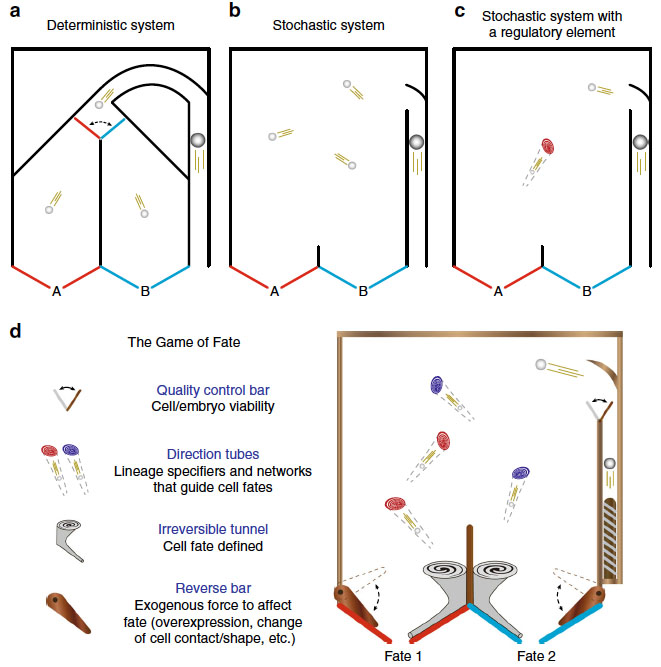
The game of fate: deterministic & stochastic factors
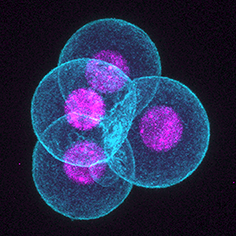

Symmetry-breaking in mammalian early embryo: when and how?
The concept of 'RNA Code' and how to decode it with advanced tools to promote precision medicine
Exploring the expanding world of small non-coding RNAs and their disease associations with new tools
Beyond RNAi: aptamer-like function of small RNAs
Small RNA structure and modification in cell function



